# Option 1: tidytuesdayR package
## install.packages("tidytuesdayR")
library(tidyverse)Refugees Analysis
data prep
read data
tuesdata <- tidytuesdayR::tt_load("2023-08-22")
data_raw <- tuesdata |> pluck(1)skim the data
check data range missing data from origin_continent oip and hst are completely missing, ill remove them from dateset date values are from 2010 to 2022
skimr::skim(data_raw)| Name | data_raw |
| Number of rows | 64809 |
| Number of columns | 16 |
| _______________________ | |
| Column type frequency: | |
| character | 6 |
| numeric | 10 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| coo_name | 0 | 1 | 4 | 52 | 0 | 210 | 0 |
| coo | 0 | 1 | 3 | 3 | 0 | 210 | 0 |
| coo_iso | 0 | 1 | 3 | 3 | 0 | 210 | 0 |
| coa_name | 0 | 1 | 4 | 52 | 0 | 189 | 0 |
| coa | 0 | 1 | 3 | 3 | 0 | 189 | 0 |
| coa_iso | 0 | 1 | 3 | 3 | 0 | 189 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| year | 0 | 1.00 | 2016.39 | 3.72 | 2010 | 2013.00 | 2017 | 2020 | 2022 | ▆▅▇▅▇ |
| refugees | 0 | 1.00 | 3440.01 | 55255.97 | 0 | 5.00 | 12 | 88 | 3737369 | ▇▁▁▁▁ |
| asylum_seekers | 0 | 1.00 | 564.17 | 7455.73 | 0 | 0.00 | 8 | 57 | 940668 | ▇▁▁▁▁ |
| returned_refugees | 0 | 1.00 | 73.48 | 2460.28 | 0 | 0.00 | 0 | 0 | 381275 | ▇▁▁▁▁ |
| idps | 0 | 1.00 | 7088.70 | 163174.63 | 0 | 0.00 | 0 | 0 | 8252788 | ▇▁▁▁▁ |
| returned_idps | 0 | 1.00 | 706.16 | 23654.24 | 0 | 0.00 | 0 | 0 | 2134349 | ▇▁▁▁▁ |
| stateless | 0 | 1.00 | 756.63 | 19980.22 | 0 | 0.00 | 0 | 0 | 955399 | ▇▁▁▁▁ |
| ooc | 0 | 1.00 | 437.50 | 20182.68 | 0 | 0.00 | 0 | 0 | 3206577 | ▇▁▁▁▁ |
| oip | 64709 | 0.00 | 196611.16 | 419586.96 | 5 | 8218.25 | 23165 | 164760 | 2453862 | ▇▁▁▁▁ |
| hst | 58845 | 0.09 | 6264.66 | 210090.46 | 0 | 0.00 | 0 | 0 | 15209720 | ▇▁▁▁▁ |
continents data
countries <- read_csv("https://gist.githubusercontent.com/fogonwater/bc2b98baeb2aa16b5e6fbc1cf3d7d545/raw/6fd2951260d8f171181a45d2f09ee8b2c7767330/countries.csv")
countries_less <- countries |> select(country_code3, continent_name)enrich
df <- data_raw |>
select(!c(oip, hst)) |>
mutate(year = as_date(parse_date_time(year, "%Y")), .before = 1) |>
rename(
origin_country = coo_name,
destination_country = coa_name,
others_of_concern = ooc
) |>
left_join(countries_less, by = join_by("coo_iso" == "country_code3")) |>
left_join(countries_less, by = join_by("coa_iso" == "country_code3")) |>
rename(
origin_continent = continent_name.x,
destination_continent = continent_name.y
)
df |> glimpse()Rows: 64,809
Columns: 16
$ year <date> 2010-01-01, 2010-01-01, 2010-01-01, 2010-01-01,…
$ origin_country <chr> "Afghanistan", "Iran (Islamic Rep. of)", "Iraq",…
$ coo <chr> "AFG", "IRN", "IRQ", "PAK", "ARE", "CHI", "GAZ",…
$ coo_iso <chr> "AFG", "IRN", "IRQ", "PAK", "EGY", "CHN", "PSE",…
$ destination_country <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Af…
$ coa <chr> "AFG", "AFG", "AFG", "AFG", "ALB", "ALB", "ALB",…
$ coa_iso <chr> "AFG", "AFG", "AFG", "AFG", "ALB", "ALB", "ALB",…
$ refugees <dbl> 0, 30, 6, 6398, 5, 6, 5, 5, 49, 5, 5, 0, 0, 6, 6…
$ asylum_seekers <dbl> 0, 21, 0, 9, 0, 0, 0, 0, 20, 0, 0, 5, 10, 92, 5,…
$ returned_refugees <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ idps <dbl> 351907, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ returned_idps <dbl> 3366, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ stateless <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ others_of_concern <dbl> 838250, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ origin_continent <chr> "Asia", "Asia", "Asia", "Asia", "Africa", "Asia"…
$ destination_continent <chr> "Asia", "Asia", "Asia", "Asia", "Europe", "Europ…data to long format
group_to_long <- function(.data, ...) {
.data |>
group_by(year, ...) |>
summarise(across(where(is.double), \(x) sum(x))) |>
pivot_longer(refugees:others_of_concern)
}
refugees_long <- group_to_long(df)
refugees_long_origin <- group_to_long(df, origin_continent)
refugees_long_destination <- group_to_long(df, destination_continent)get colorspace
library(colorspace)
qualitative_hcl(6)[1] "#E16A86" "#B88A00" "#50A315" "#00AD9A" "#009ADE" "#C86DD7"high level analysis
notes
origin and destinations countries which general direction of refugees movement. refugees group are split to different sub classes, like asylum seekers, internally displaced persons and stateless person refugee and internally displaced person also have returned level
focus
check which refugees group gives interesting analysis layer
global level
scale_y_mil <- function() {
scale_y_continuous(labels = scales::label_number(scale = 1 / 1000000, big.mark = " ", suffix = "M"))
}
scale_x_yrs <- function() {
scale_x_date(date_labels = "%y", date_breaks = "4 years")
}
refugees_long |>
ggplot(aes(year, value, fill = name)) +
geom_col() +
scale_y_mil() +
scale_x_yrs() +
labs(
title = "has there been a change in refugees on global level?",
subtitle = "2010 - 2023"
) +
scale_fill_discrete_qualitative()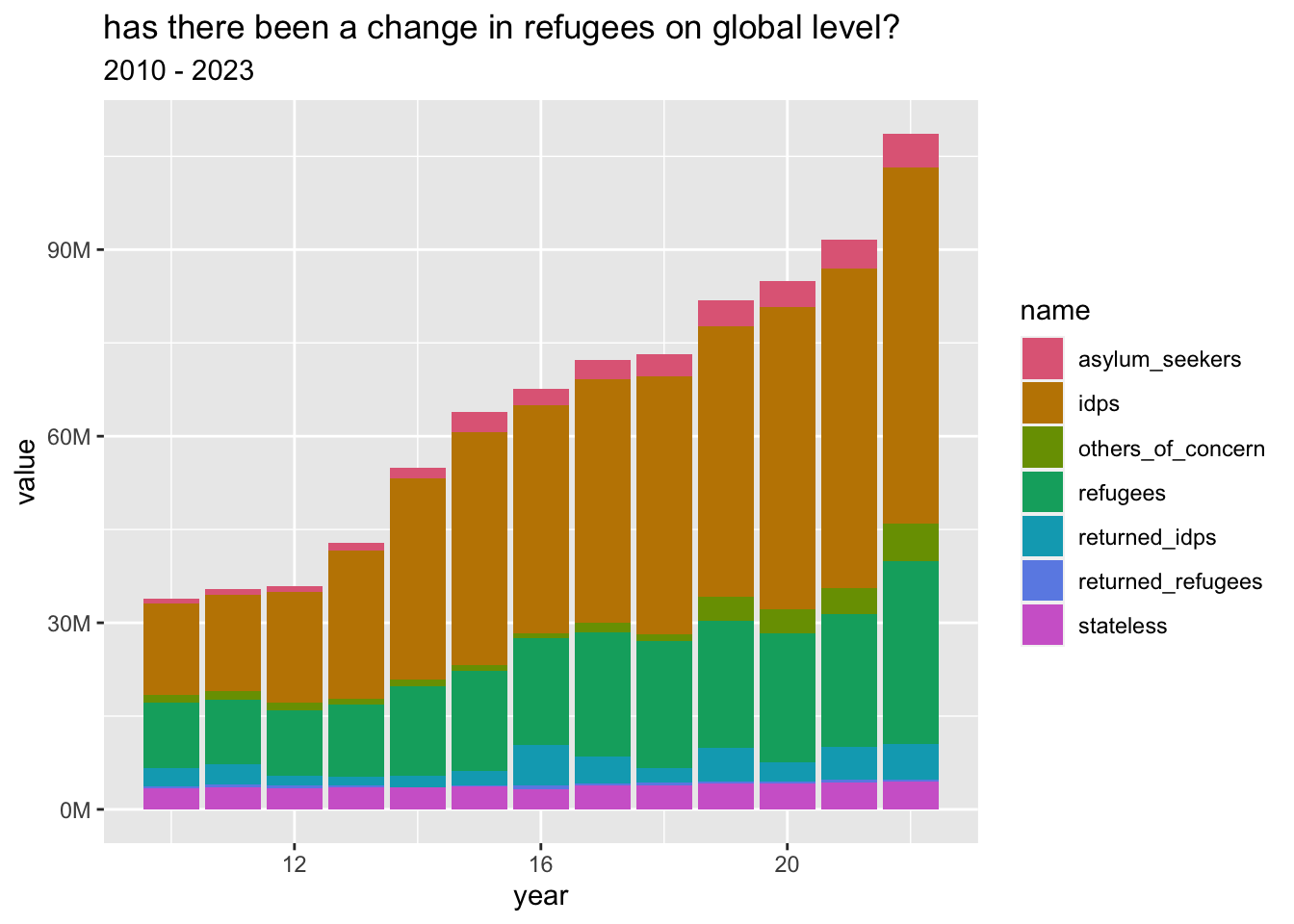
continent level
plot_continents <- function(.data, .title = "value", .subtitle = "value2", facet_var = origin_continent, .name = name) {
.data |>
ggplot(aes(year, value, fill = {{ .name }})) +
geom_col() +
scale_x_yrs() +
scale_y_mil() +
labs(
title = .title,
subtitle = .subtitle
) +
scale_fill_discrete_qualitative() +
facet_grid(cols = vars({{ facet_var }}))
}
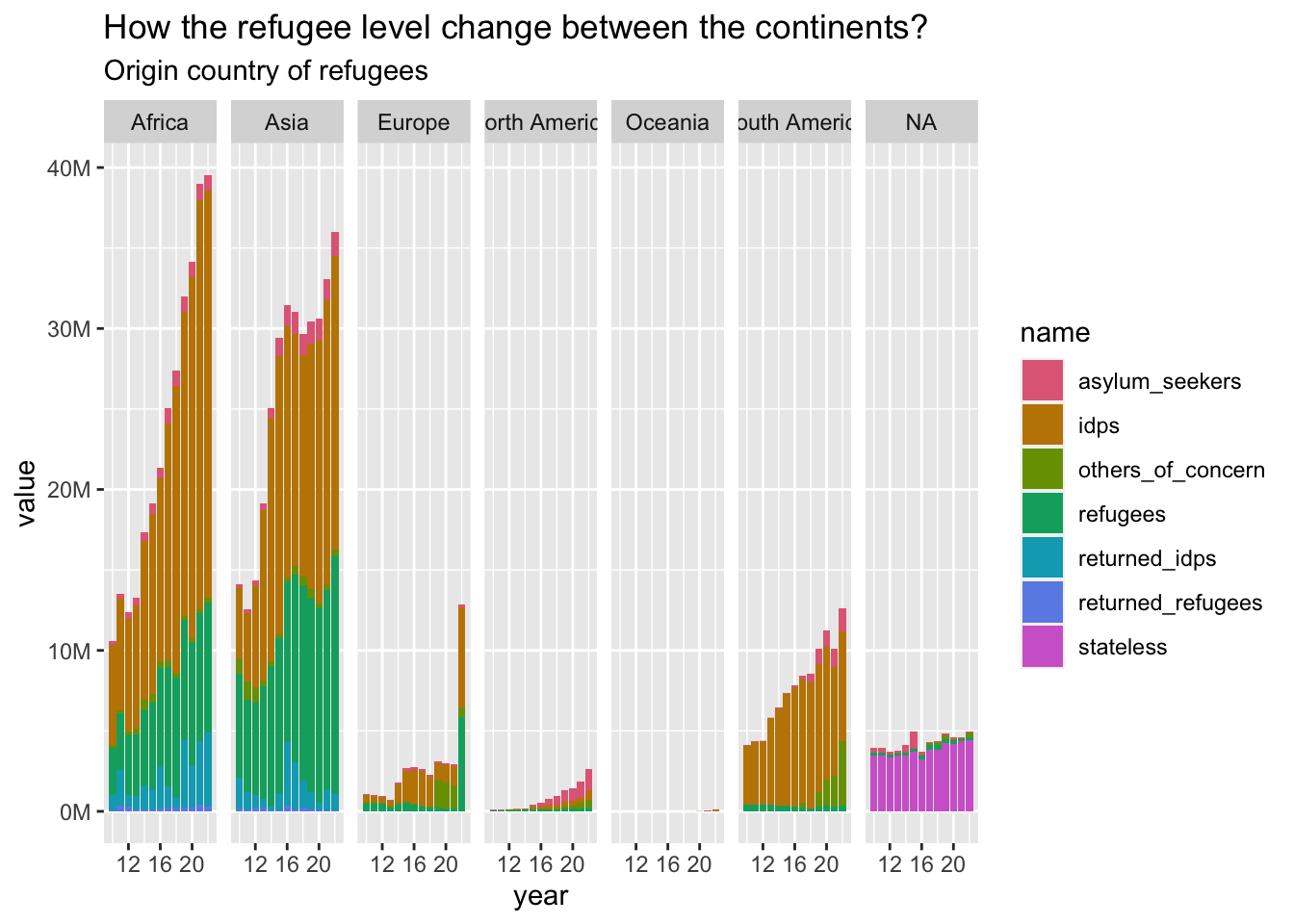
plot_continents(
refugees_long_origin, "How the refugee level change between the continents?",
"Origin country of refugees"
)
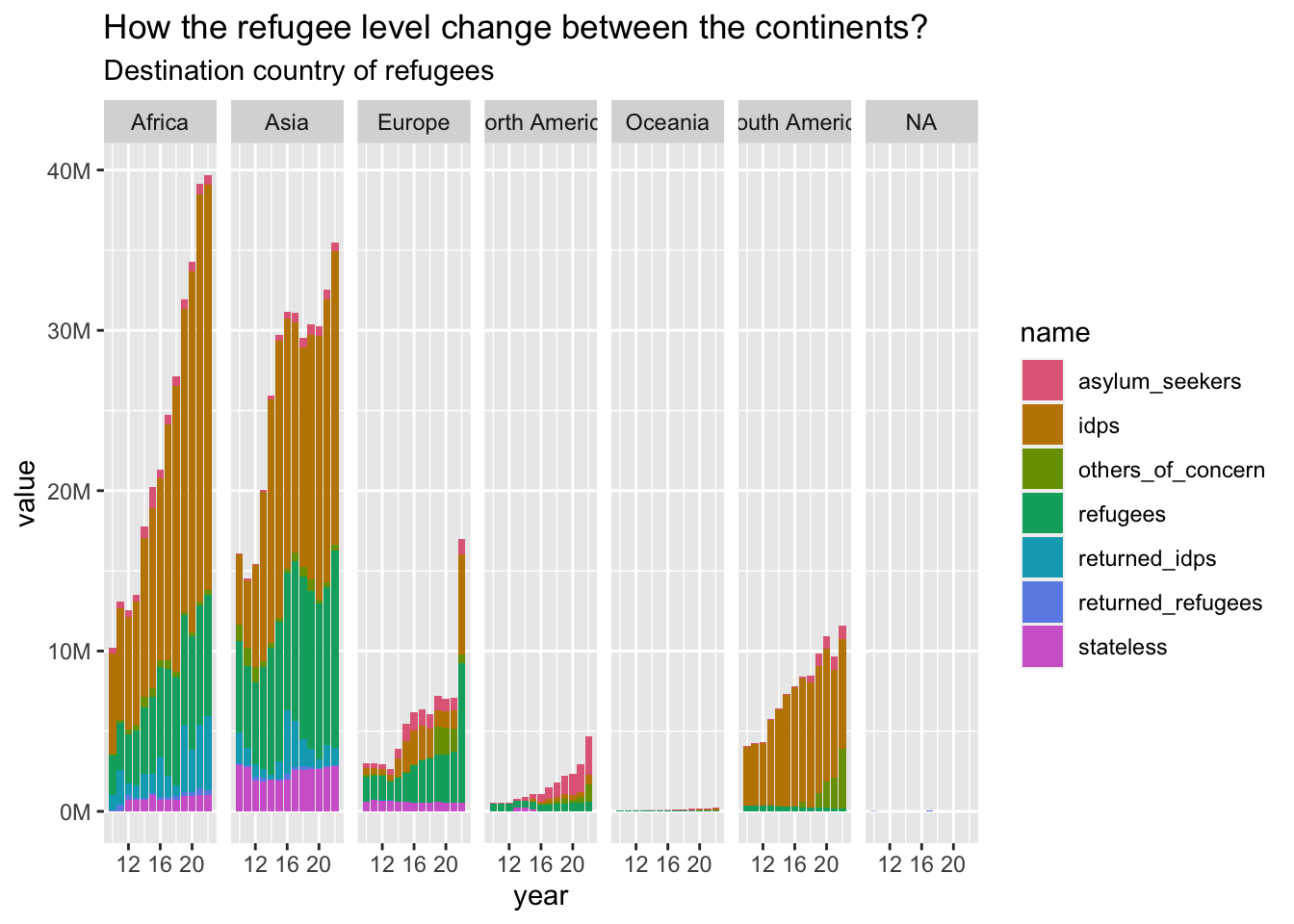
plot_continents(
refugees_long_destination, "How the refugee level change between the continents?",
"Destination country of refugees", destination_continent
)
refugees between the continets
there seems to be intra continent refugees, next ill check how between the continets and domestic differ
df2 <- df |>
mutate(between_continents = if_else(origin_continent != destination_continent, TRUE, FALSE), .before = 1)df2 |>
filter(between_continents == TRUE) |>
group_to_long(destination_continent) |>
plot_continents("what is the destination of Between continents refugees", "", destination_continent)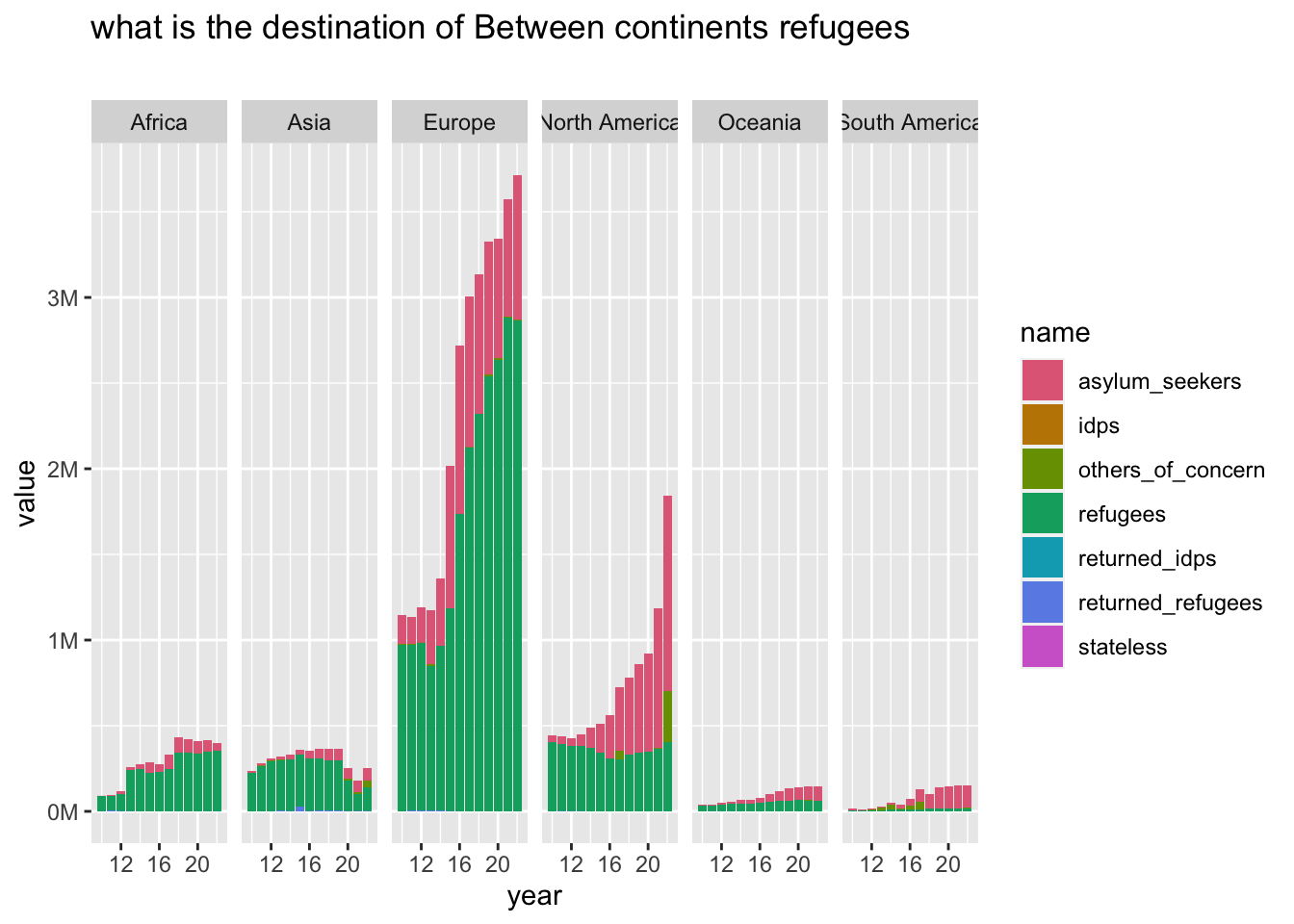
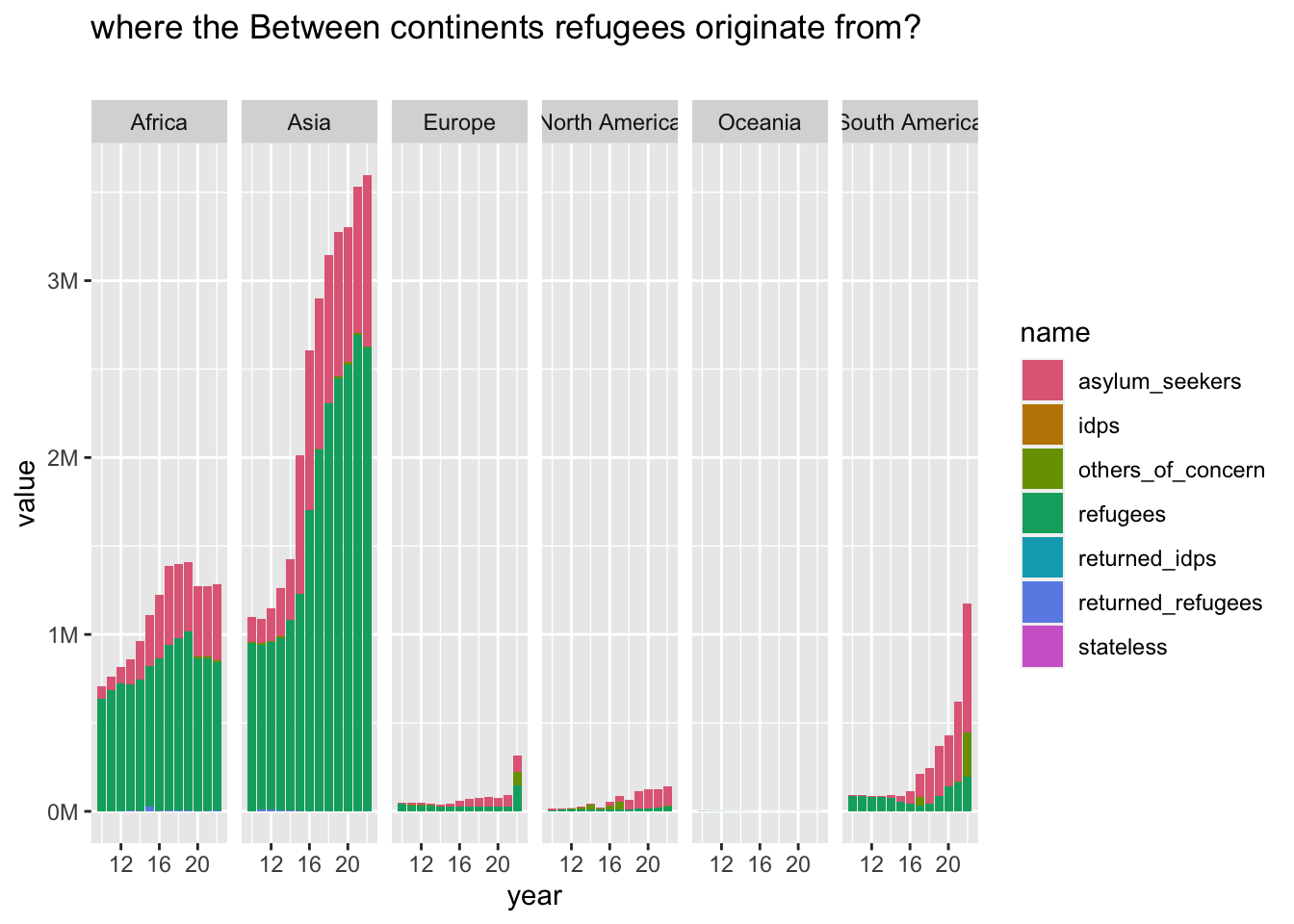
df2 |>
filter(between_continents == TRUE) |>
group_to_long(origin_continent) |>
plot_continents(
"where the Between continents refugees originate from?", "",
origin_continent
)
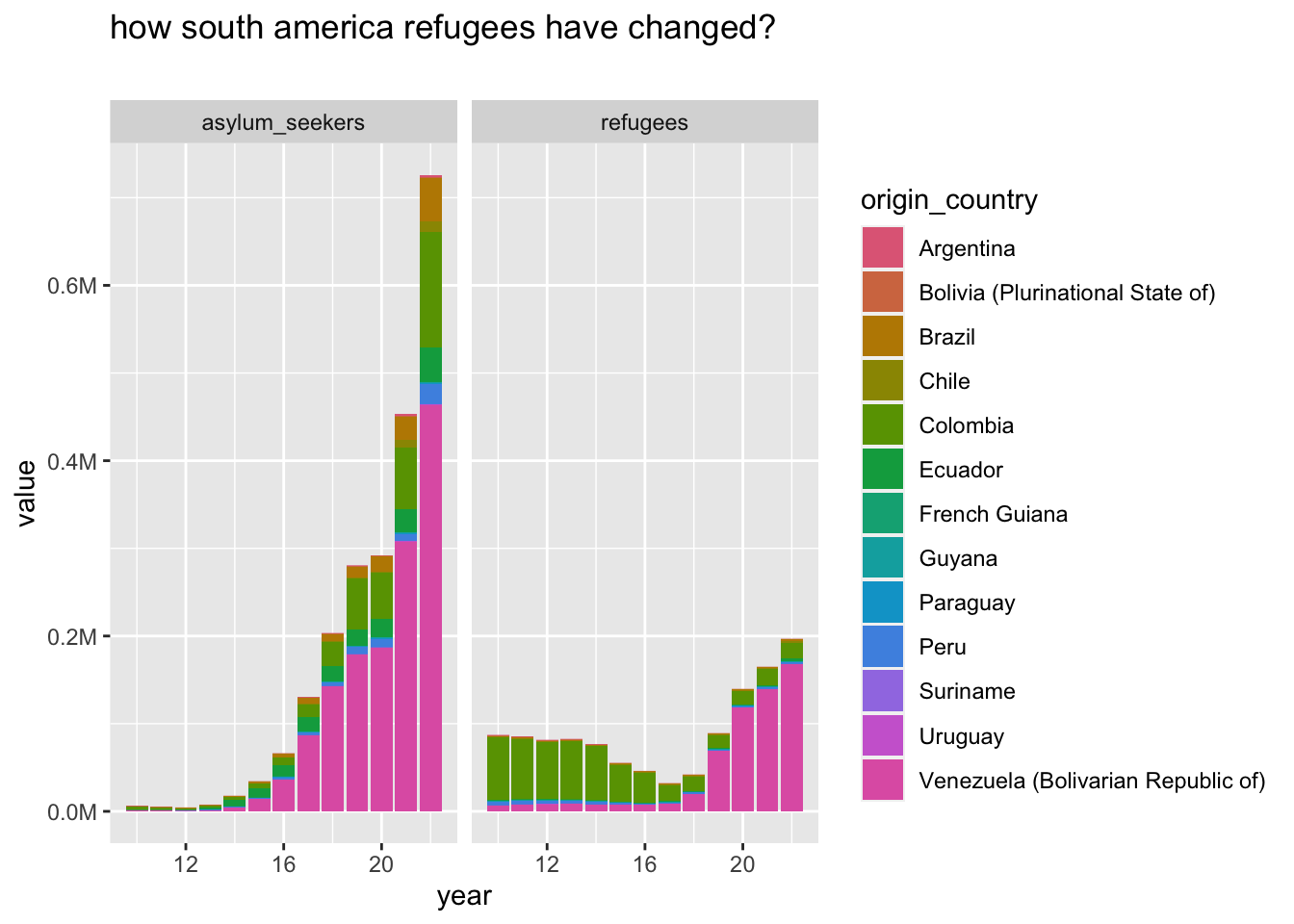
deep dive south america, between continets
south america has interesting change, and I’m not familiar with the continent. I’ll try to check if data gives interesting elements
southAmerica <- df2 |>
filter(origin_continent == "South America", between_continents == TRUE)southAmericaSummarized <- southAmerica |>
select(-year) |>
group_by(origin_country) |>
summarise(across(where(is.double), list(sum = sum)))
southAmericaSummarized |>
gt::gt()| origin_country | refugees_sum | asylum_seekers_sum | returned_refugees_sum | idps_sum | returned_idps_sum | stateless_sum | others_of_concern_sum |
|---|---|---|---|---|---|---|---|
| Argentina | 3045 | 6788 | 0 | 0 | 0 | 0 | 20 |
| Bolivia (Plurinational State of) | 4767 | 6888 | 0 | 0 | 0 | 0 | 0 |
| Brazil | 16218 | 127858 | 0 | 0 | 0 | 0 | 3574 |
| Chile | 8302 | 25238 | 0 | 0 | 0 | 0 | 0 |
| Colombia | 510459 | 383772 | 40 | 0 | 0 | 0 | 45253 |
| Ecuador | 15796 | 176299 | 0 | 0 | 0 | 0 | 24525 |
| French Guiana | 11 | 238 | 0 | 0 | 0 | 0 | 0 |
| Guyana | 6193 | 4578 | 0 | 0 | 0 | 0 | 0 |
| Paraguay | 1017 | 3537 | 0 | 0 | 0 | 0 | 0 |
| Peru | 32663 | 64606 | 0 | 0 | 0 | 0 | 0 |
| Suriname | 250 | 635 | 0 | 0 | 0 | 0 | 0 |
| Uruguay | 1028 | 2470 | 0 | 0 | 0 | 0 | 0 |
| Venezuela (Bolivarian Republic of) | 577459 | 1422868 | 5 | 0 | 0 | 0 | 230430 |
select refugees and asylum seekers since the other refugree classes are mostly zeros
southAmerica |>
group_to_long(origin_country) |>
filter(name %in% c("refugees", "asylum_seekers")) |>
plot_continents("how south america refugees have changed?", "", facet_var = name, .name = origin_country)